Taxonomy, tagging, and topics oh my
2024-10-20:
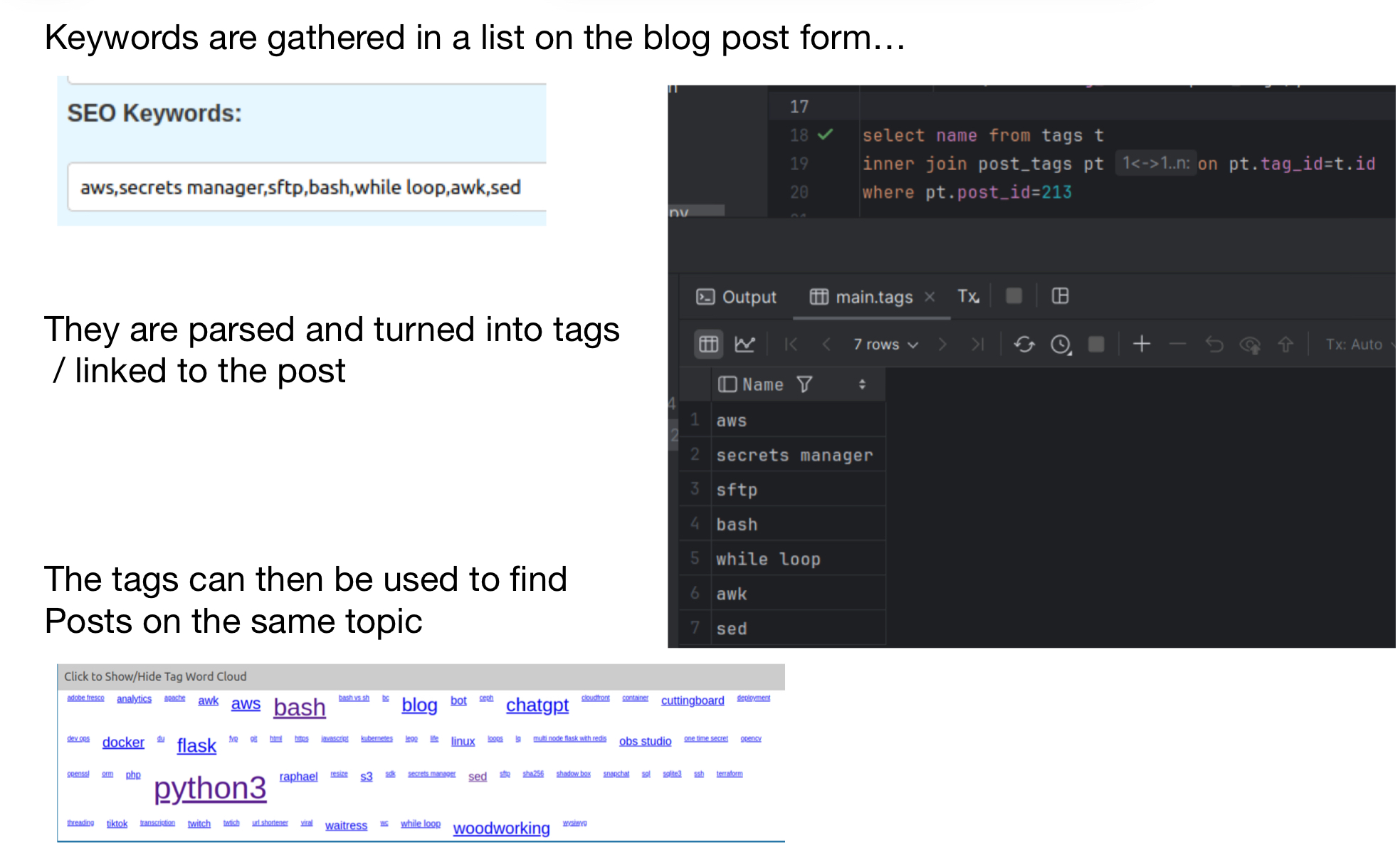
This weekend I decided that I wanted an easier way to look up things I had written by the topics I’m writing about. I’m a fan of normalized data so I started by creating a couple tables. One for the tags aka topics that I’ve written about, and one for a link between my posts and the topics. I was day-dreaming the user interface for these topics and had started muddling through creating some endpoints when it occurred to me that I already had topics that I had applied to the posts in the way of a string of topics used for SEO (search engine optimization). With this end mind I decided to write a script whose job would be to loop through all of my blog posts and process the tags/topics one by one. This script would create the tag/topic if it didn’t exist and then create the link between the posts and the tags. After debugging that process it occurred to me that a similar process could be used so that the user interface for maintaining the tabs could be the existing blog post form, as is, without any changes. This could be accomplished by taking the existing seo data, spiting it by commas and then processing the topics/tags one at a time. All that was left was code to clean up anything that was removed when editing posts.

Dynamic code generation to save time
8/11/2024:
This weekend, I spent some time trying to look for patterns in what should be random file names. I started the process by writing some Python to read a text file with the results of an s3 ls command, break the file name into pieces for each character of the file name, and insert it into an SQLite3 database. I used some new-to-me bash techniques for code generation using a command line for loop. Along with an older sql creation by sql. This with the newer execute a command for each file line and I was in business.
#add column numbers
#for f = 1 to 50
#add 50 columns named f1 to f50
for f in {1..50}
do
sqlite3 fileanalysis.db "alter table files add column f$f text;"
done
#loop for f = 1 to 48:
#loop for dynamic sql insert statements in mass
for f in {1..48}
do
sqlite3 control.db "insert into file_name_position_charecter_counts (f,charecter) select distinct 'f$f', f$f from files order by f$f";
done
#loop through sql generating sql and run against our sqlite3 database.
while IFS= read -r sql
do
sqlite3 control.db "$sql" > sqllog.txt
done < control_counts.tsv
--Create update statements to do charecter counts
select 'update file_name_position_charecter_counts set count=(select count(*) from files where '
|| c.f || '=''' || c.charecter || ''') where id = ' || cast(c.id as text)
from file_name_position_charecter_counts c;